Exploring pollution in a pandemic
You may remember the stories in the early pandemic months of 2020 telling us how persistent air pollution was clearing up as cities and countries went into lockdown. We heard about views of the Himalayas. Similarly, Los Angeles, notorious for air pollution, recorded its cleanest air in years. But things didn’t stay that way for long. I wanted to take a look at the air quality data for New York City, where I live, to see what insight I could glean. It was a strange summer in 2020, with a rise in car traffic, lots of grilling, lots of fireworks, and innumerable other small changes to the way that the city typically emits pollutants into the atmosphere.
I started by taking a trip to the EPA data website. The EPA operates air quality monitoring stations throughout the US, including multiple stations in NYC. They provide daily air quality data from these stations.
I’m going to walk through the analyses I did here at the end of this post, but all the data I gathered and the code to run these analyses (in R) can be found in this Github repository.
The short story is that things are weird.
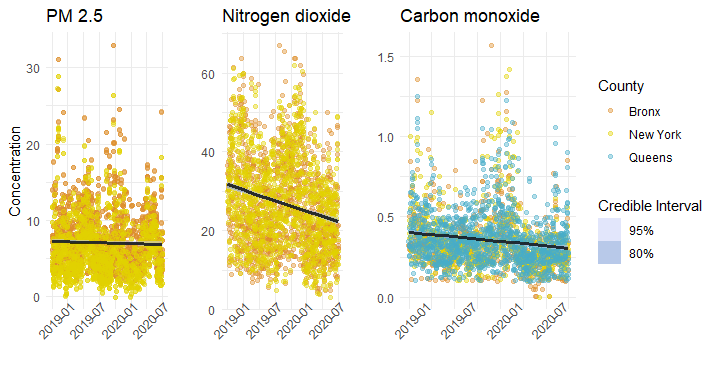
Joy plot of pollutant concentrations by month. The distribution for each month describes the totality of daily measurements for that month. Light-colored curves are months during the pandemic. Darker curves are pre-pandemic.
At first glance, we can pick out some interesting insights from our joy plots.
Every summer we see an increase in PM 2.5 between July and August, a pattern that is not really mirrored in Carbon monoxide. There are a few possible explanations here, but one contributor might be fireworks. This seems especially reasonable given the large increase in summer 2020, when many industries were operating at lower capacity, but when fireworks were rampant in the city. In general however, it seems that there has been a slight easing of fine particulate pollution over the course of the pandemic. (Here the pandemic is colored with a lighter blue).
Nitrogen dioxide, which is a product of fossil fuel emissions, has been relatively steady over time, with a fairly predictable meander upwards each spring, potentially due to warming temperatures and atmospheric effects.
Of interest to our question here, of how the shutdown of the city has affected air quality, is the hard decline of Carbon monoxide in the early months of the pandemic. In April through September, CO levels appear to have been at some of their lowest concentrations in the last 5 years.
One way to think about this problem is to look at the concentrations in the month of April, since NYC was largely shuttered then. We can compare April 2020 to other Aprils.
The EPA data csvs come with a variety of data columns:
In these violin plots, the red plot is the pandemic April - 2020.
When viewed this way - comparing Aprils in different years, we can see that April 2020 showed some difference, but it’s not clear in each case whether that difference is substantially different enough from normal that we can think about attributing causation to it.
Below I walk through the full analysis, including some basic models and some data exploration.
In this example, I’m going to walk through an approach to looking at the question of whether the COVID shutdown in New York City had a measurable effect on air quality. In particular, I’ll look at 3 common pollutants - Carbon monoxide, Nitrogen dioxide, and fine particulate matter (PM 2.5) as recorded by EPA air quality monitoring stations.
First we’ll do some setup. Here I’ll load packages to help with data munging, visualization, and analysis.
# For reproducibility set.seed(88) # Load packages - Munging library(dplyr) library(lubridate) # Visualization library(ggplot2) library(ggjoy) library(wesanderson) library(cowplot) library(tidybayes) # Analysis library(R2jags) library(rstanarm) library(modelr)
We’ll import the data. EPA provides this in 1-year csv files, so we’ll have to combine them. The data are also available via the EPA API but, given the relatively small amount of data we want, in this case it’s easiest to just download individual files and combine them.
Here I’ll load in the csvs for each pollutant together, to end up with 3 data frames, one for each pollutant.
nyc_pm2_5.dat<-rbind(read.csv("data/NYNJ_PM2_5_EPA_2016.csv"), read.csv("data/NYNJ_PM2_5_EPA_2017.csv"), read.csv("data/NYNJ_PM2_5_EPA_2018.csv"), read.csv("data/NYNJ_PM2_5_EPA_2019.csv"), read.csv("data/NYNJ_PM2_5_EPA_2020.csv"), read.csv("data/NYNJ_PM2_5_EPA_2021.csv")) nyc_CO.dat<-rbind(read.csv("data/NYNJ_CO_EPA_2016.csv"), read.csv("data/NYNJ_CO_EPA_2017.csv"), read.csv("data/NYNJ_CO_EPA_2018.csv"), read.csv("data/NYNJ_CO_EPA_2019.csv"), read.csv("data/NYNJ_CO_EPA_2020.csv"), read.csv("data/NYNJ_CO_EPA_2021.csv")) nyc_NO2.dat<-rbind(read.csv("data/NYNJ_NO2_EPA_2016.csv"), read.csv("data/NYNJ_NO2_EPA_2017.csv"), read.csv("data/NYNJ_NO2_EPA_2018.csv"), read.csv("data/NYNJ_NO2_EPA_2019.csv"), read.csv("data/NYNJ_NO2_EPA_2020.csv"), read.csv("data/NYNJ_NO2_EPA_2021.csv")) names(nyc_pm2_5.dat)
## [1] "Date" "Source" ## [3] "Site.ID" "POC" ## [5] "Daily.Mean.PM2.5.Concentration" "UNITS" ## [7] "DAILY_AQI_VALUE" "Site.Name" ## [9] "DAILY_OBS_COUNT" "PERCENT_COMPLETE" ## [11] "AQS_PARAMETER_CODE" "AQS_PARAMETER_DESC" ## [13] "CBSA_CODE" "CBSA_NAME" ## [15] "STATE_CODE" "STATE" ## [17] "COUNTY_CODE" "COUNTY" ## [19] "SITE_LATITUDE" "SITE_LONGITUDE"
names(nyc_CO.dat)
## [1] "Date" "Source" ## [3] "Site.ID" "POC" ## [5] "Daily.Max.8.hour.CO.Concentration" "UNITS" ## [7] "DAILY_AQI_VALUE" "Site.Name" ## [9] "DAILY_OBS_COUNT" "PERCENT_COMPLETE" ## [11] "AQS_PARAMETER_CODE" "AQS_PARAMETER_DESC" ## [13] "CBSA_CODE" "CBSA_NAME" ## [15] "STATE_CODE" "STATE" ## [17] "COUNTY_CODE" "COUNTY" ## [19] "SITE_LATITUDE" "SITE_LONGITUDE"
names(nyc_NO2.dat)
## [1] "Date" "Source" ## [3] "Site.ID" "POC" ## [5] "Daily.Max.1.hour.NO2.Concentration" "UNITS" ## [7] "DAILY_AQI_VALUE" "Site.Name" ## [9] "DAILY_OBS_COUNT" "PERCENT_COMPLETE" ## [11] "AQS_PARAMETER_CODE" "AQS_PARAMETER_DESC" ## [13] "CBSA_CODE" "CBSA_NAME" ## [15] "STATE_CODE" "STATE" ## [17] "COUNTY_CODE" "COUNTY" ## [19] "SITE_LATITUDE" "SITE_LONGITUDE"
Data Munging
The files we get are pollutant-specific, but fortunately nearly all the column names are the same - the only difference is the name of the measurement. They’re all some form of concentration, and the precise units are unimportant to us here. So we can simply relabel them each as “Concentration.” In this way, our columns are shared across the datasets, and we can fuse them. There are a few other tasks to accomplish. We’ll want some easier variables to work with than the full date, like the year and month, so we’ll create those here. We’ll also create a column called “Pandemic” which is a factor that describes whether the measurement was taken pre-pandemic (Normal) or during (Pandemic). We can also jettison the data for areas outside of NYC at this point. These might be interesting, but our particular question relates to NYC.
# Each data frame has a different column name for the measurement. We'll standardize nyc_pm2_5.dat<-nyc_pm2_5.dat %>% rename(Concentration = Daily.Mean.PM2.5.Concentration) nyc_CO.dat<-nyc_CO.dat %>% rename(Concentration=Daily.Max.8.hour.CO.Concentration) nyc_NO2.dat<-nyc_NO2.dat %>% rename(Concentration=Daily.Max.1.hour.NO2.Concentration) # now we can put them together nyc.dat<-dplyr::bind_rows(nyc_pm2_5.dat, nyc_CO.dat, nyc_NO2.dat) # translate date column into date objects nyc.dat$Date<-lubridate::mdy(nyc.dat$Date) # Date munging nyc.dat$Year<-lubridate::year(nyc.dat$Date) # Add year column nyc.dat$Month<-lubridate::month(nyc.dat$Date) # Add month column nyc.dat$Day<-lubridate::yday(nyc.dat$Date) # Add day-of-year column # It would be nice to plot everything chronologically, but still have factors as months. # We can do this by creating a column that is a numeric representation of months nyc.dat$RunningMonth<-as.numeric(nyc.dat$Year)+as.numeric(nyc.dat$Month)/13 # Then we make a more readable format nyc.dat$RunningtextMonth<-paste(month(nyc.dat$Date, label = TRUE), nyc.dat$Year) # And use the numeric format to re-order the readable format so we can plot with it nyc.dat$RunningtextMonth<-reorder(nyc.dat$RunningtextMonth, nyc.dat$RunningMonth) # Add a column to delimit the start of Covid in NYC nyc.dat$Pandemic<-sapply(nyc.dat$RunningMonth, FUN=function(x) ifelse(x < 2020.3, "Normal", "Pandemic")) # We'll make a little vector of the pollutant names to help us along pol_types<-unique(nyc.dat$AQS_PARAMETER_DESC) # The data are for the wider NYC area including NJ and Long Island # We'll pull only the relevant counties nyc.dat<-dplyr::filter(nyc.dat, COUNTY %in% c("Kings", "Queens", "Bronx", "New York", "Richmond"), AQS_PARAMETER_DESC %in% pol_types[c(1,3:4)])
Preliminary Visualization
We can visualize the data with a simple scatterplot over time to see how concentration changes. We’ll set colors based on borough/county in NYC. But it turns out that this is misleading, since a dip in 2020 is actually due to a lack of readings for some pollutants.
ggplot(data=nyc.dat[nyc.dat$Year > 2017 & nyc.dat$AQS_PARAMETER_DESC == "PM2.5 - Local Conditions",], aes(x=Day, y=Concentration))+ geom_jitter(aes(color=COUNTY), alpha = 0.2, height = 0.03)+ theme_minimal()+ labs(x = "Day of the Year")+ ggtitle("PM 2.5")+ facet_wrap(~Year)
Joy Plots
Joy plots can help us understand the change over time, visualized in a really rich plot that looks like a ridgeline. I’ll make a joy plot for each pollutant and we can look at them together over time.
# PM 2.5 Plot
joy.pm25<-ggplot(nyc.dat[nyc.dat$AQS_PARAMETER_DESC == pol_types[1],],
aes(x = Concentration,
y = factor(RunningtextMonth)))+
geom_joy(scale = 3,
alpha = 0.95,
size = 1,
aes(fill = Pandemic,
color = Pandemic))+
labs(x = "")+
lims(x = c(-5,25))+
ggtitle("PM 2.5")+
scale_color_manual(values = wes_palette("Zissou1")[c(2,1)])+
scale_fill_manual(values = wes_palette("Zissou1")[c(1,2)])+
scale_y_discrete(breaks = levels(nyc.dat$RunningtextMonth)[c(T,F)])+
theme_minimal()+
theme(legend.position = "None",
axis.title.y = element_blank())
# Carbon monoxide plot
joy.CO<-ggplot(nyc.dat[nyc.dat$AQS_PARAMETER_DESC == pol_types[3],],
aes(x = Concentration,
y = factor(RunningtextMonth)))+
geom_joy(scale = 3,
alpha = 0.95,
size = 1,
aes(fill = Pandemic,
color = Pandemic))+
labs(x = "Concentration", y = "")+
lims(x = c(0,1))+
ggtitle(pol_types[3])+
scale_color_manual(values = wes_palette("Zissou1")[c(2,1)])+
scale_fill_manual(values = wes_palette("Zissou1")[c(1,2)])+
theme_minimal()+
theme(legend.position = "None",
axis.text.y = element_blank(),
axis.title.y = element_blank())
# Nitrogen dioxide plot
joy.NO2<-ggplot(nyc.dat[nyc.dat$AQS_PARAMETER_DESC == pol_types[4],],
aes(x = Concentration,
y = factor(RunningtextMonth)))+
geom_joy(scale = 3,
alpha = 0.95,
size = 1,
aes(fill = Pandemic,
color = Pandemic))+
labs(x = "", y = "")+
lims(x = c(-5,75))+
ggtitle("Nitrogen dioxide")+
scale_color_manual(values = wes_palette("Zissou1")[c(2,1)])+
scale_fill_manual( values = wes_palette("Zissou1")[c(1,2)])+
theme_minimal()+
theme(legend.position = "None",
axis.title.y = element_blank(),
axis.text.y = element_blank())
# Combine plots
cowplot::plot_grid(joy.pm25,
joy.CO,
joy.NO2,
ncol = 3,
rel_widths = c(0.38,0.31,0.31))
At first glance, we can pick out some interesting insights from our joy plots.
Every summer we see an increase in PM 2.5 between July and August, a pattern that is not really mirrored in Carbon monoxide. There are a few possible explanations here, but one contributor might be fireworks. This seems especially reasonable given the large increase in summer 2020, when many industries were operating at lower capacity, but when fireworks were rampant in the city. In general however, it seems that there has been a slight easing of fine particulate pollution over the course of the pandemic. (Here the pandemic is colored with a lighter blue).
Nitrogen dioxide, which is a product of fossil fuel emissions, has been relatively steady over time, with a fairly predictable meander upwards each spring, potentially due to warming temperatures and atmospheric effects.
Of interest to our question here, of how the shutdown of the city has affected air quality, is the hard decline of Carbon monoxide in the early months of the pandemic. In April through September, CO levels appear to have been at some of their lowest concentrations in the last 5 years.
To gain an overall sense of the change, we can use the EPA’s Air Quality Index statistic in another joy plot:
ggplot(data = nyc.dat,
aes(x = DAILY_AQI_VALUE,
y = RunningtextMonth))+
geom_joy(scale = 4,
alpha = 0.95,
size = 1,
aes(fill = Pandemic,
color = Pandemic))+
lims(x = c(-10, 90))+
labs(x = "Air Quality Index")+
theme_minimal()+
theme(axis.title.y = element_blank(),
legend.position = "None")+
ggtitle("EPA Air Quality - NYC")+
scale_color_manual(values = wes_palette("Zissou1")[c(2,1)])+
scale_fill_manual( values = wes_palette("Zissou1")[c(1,2)])+
scale_y_discrete(breaks = levels(nyc.dat$RunningtextMonth)[c(T,F)])
The AQI, calculated by the EPA, is not an ideal statistic to be visualized this way, since it operates on a discrete scale. In our joy plot, you can see the bump at low levels get smoothed.
ggplot(data = nyc.dat, aes(x = Date, y = DAILY_AQI_VALUE))+ geom_jitter(aes(color = Pandemic), height = 0.25, alpha = 0.2)+ theme_minimal()+ labs(y = "Daily Air Quality Index")
When we just plot out the AQI as a scatterplot, we see a big dip right at the beginning of the “Pause” in NYC. But it’s not clear if this is driven by actual changes in air quality, or missing data.
ggplot(data = nyc.dat[nyc.dat$Year > 2018,], aes(x = Date, y = DAILY_AQI_VALUE))+ geom_jitter(aes(color = Pandemic), height = 0.25, alpha = 0.2)+ theme_minimal()+ theme(axis.text.x = element_text(angle=35, hjust = 1))+ labs(y = "Daily Air Quality Index", x = "")+ facet_wrap(~COUNTY)
When we compare boroughs side-by-side for 2019 and 2020, we can see where the data are sparse, and where they remain good. Manhattan for example (New York County) seems less dense. But is that just because the air quality dramatically improved as office workers stayed home, and thus all the points are clustered at the bottom?
ggplot(data=nyc.dat[nyc.dat$Year == 2020,], aes(month(Month, label = T)))+ geom_bar(aes(fill = COUNTY))+ theme_minimal()+ theme(axis.text.x = element_text(angle = 45))+ ggtitle("2020")+ labs(x = "Month", y = "No. of Measurements")+ scale_fill_manual(values = wes_palette("Darjeeling1", type = "discrete")[c(2,3,1,4,5)])+ facet_wrap(~AQS_PARAMETER_DESC)
Broken down a little more explicitly, we see that there are no measurements of Nitrogen dioxide anywhere but the Bronx and Queens in 2020. For local PM 2.5, at the beginning of the shutdown, no measurements come from Staten Island (Richmond), Manhattan (New York), or Brooklyn (Kings), though all return by August. For CO, no measurements come from Staten Island and Brooklyn, but other boroughs have consistent measurements.
So if our interest is in understanding pandemic-related changes in air quality, we should stick to those boroughs with consistent data. The Bronx and Queens are both reliable here throughout, and Manhattan can help us understand Carbon monoxide as well.
Modelling air quality during covid
Given the degree of change in behavior of residents in late March through early May, it seems simplest to focus on that period to ask our most basic question of whether the pandemic had a meaningful effect on these pollutants.
So we’ll start by working up a simple linear model, implemented here in a Bayesian framework with Stan, comparing the concentration of pollutants over the course of 2019 through mid-2020.
# Build out our linear model for each pollutant CO_lm.stan<-stan_lm(data = filter(nyc.dat, COUNTY %in% c("Bronx", "New York", "Queens"), AQS_PARAMETER_DESC %in% "Carbon monoxide", RunningMonth > 2019, RunningMonth < 2020.7), Concentration~Date, prior = R2(0.5, 'mean')) NO2_lm.stan<-stan_lm(data = filter(nyc.dat, COUNTY %in% c("Bronx", "Queens"), AQS_PARAMETER_DESC %in% "Nitrogen dioxide (NO2)", RunningMonth > 2019, RunningMonth < 2020.7), Concentration~Date, prior = R2(0.5, 'mean')) PM2_5_lm.stan<-stan_lm(data = filter(nyc.dat, COUNTY %in% c("Bronx", "Queens"), AQS_PARAMETER_DESC %in% "PM2.5 - Local Conditions", RunningMonth > 2019, RunningMonth < 2020.7), Concentration~Date, prior = R2(0.5, 'mean')) # Plot posterior draws and credible intervals on the data CO_lm.plot<-filter(nyc.dat, COUNTY %in% c("Bronx", "New York", "Queens"), AQS_PARAMETER_DESC %in% "Carbon monoxide", RunningMonth > 2019, RunningMonth < 2020.7) %>% data_grid(Date = seq_range(Date, n = 10), Concentration = seq_range(Concentration,by = 0.1)) %>% add_fitted_draws(CO_lm.stan, n = 100) %>% ggplot(aes(x = Date, y = Concentration))+ geom_jitter(data = filter(nyc.dat, COUNTY %in% c("Bronx", "New York", "Queens"), AQS_PARAMETER_DESC %in% "Carbon monoxide", Year > 2018, RunningMonth < 2020.7), alpha = 0.4, height = 0.1, aes(color = COUNTY))+ stat_lineribbon(aes(y = .value), .width = c(0.8, 0.95), alpha = 0.5)+ theme_minimal()+ theme(axis.text.x = element_text(angle = 45))+ ggtitle("Carbon monoxide")+ scale_fill_manual(values = wes_palette("GrandBudapest2")[c(2,4)], name = "Credible Interval", labels = c("95%", "80%"))+ labs(x = "", y = "")+ scale_color_manual(name = "County", values = wes_palette("FantasticFox1")[c(1:3,5)]) NO2_lm.plot<-filter(nyc.dat, COUNTY %in% c("Bronx", "Queens"), AQS_PARAMETER_DESC %in% "Nitrogen dioxide (NO2)", RunningMonth > 2019, RunningMonth < 2020.7) %>% data_grid(Date = seq_range(Date, n = 10), Concentration = seq_range(Concentration, by = 0.1)) %>% add_fitted_draws(NO2_lm.stan, n = 100) %>% ggplot(aes(x = Date, y = Concentration))+ geom_jitter(data = filter(nyc.dat, COUNTY %in% c("Bronx", "Queens"), AQS_PARAMETER_DESC %in% "Nitrogen dioxide (NO2)", Year > 2018, RunningMonth < 2020.7), alpha = 0.4, height = 0.1, aes(color = COUNTY))+ stat_lineribbon(aes(y = .value), .width = c(0.8, 0.95), alpha = 0.5)+ theme_minimal()+ theme(legend.position = "None", axis.text.x = element_text(angle = 45))+ ggtitle("Nitrogen dioxide")+ scale_fill_manual(values = wes_palette("GrandBudapest2")[c(2,4)], name = "Credible Interval", labels = c("95%", "80%"))+ labs(x = " ", y = "")+ scale_color_manual(name = "County", values = wes_palette("FantasticFox1")[c(1:3,5)]) PM2_5_lm.plot<-filter(nyc.dat, COUNTY %in% c("Bronx", "Queens"), AQS_PARAMETER_DESC %in% "PM2.5 - Local Conditions", RunningMonth > 2019, RunningMonth < 2020.7) %>% data_grid(Date = seq_range(Date, n = 10), Concentration = seq_range(Concentration, by = 0.1)) %>% add_fitted_draws(PM2_5_lm.stan, n = 100) %>% ggplot(aes(x = Date, y = Concentration))+ geom_jitter(data = filter(nyc.dat, COUNTY %in% c("Bronx", "Queens"), AQS_PARAMETER_DESC %in% "PM2.5 - Local Conditions", Year > 2018, RunningMonth < 2020.7), alpha = 0.4, height = 0.1, aes(color = COUNTY))+ geom_jitter(data = filter(nyc.dat, COUNTY %in% c("Bronx", "Queens"), AQS_PARAMETER_DESC %in% "PM2.5 - Local Conditions", Year > 2018, RunningMonth < 2020.7), alpha = 0.4, height = 0.1, aes(color = COUNTY))+ stat_lineribbon(aes(y = .value), .width = c(0.8, 0.95), alpha = 0.5)+ theme_minimal()+ theme(legend.position = "None", axis.text.x = element_text(angle = 45))+ scale_fill_manual(values = wes_palette("GrandBudapest2")[c(2,4)], name = "Credible Interval", labels = c("95%", "80%"))+ scale_color_manual(name = "County", values = wes_palette("FantasticFox1")[c(1:3,5)])+ labs(x = "", y = "Concentration")+ ggtitle("PM 2.5") cowplot::plot_grid(PM2_5_lm.plot, NO2_lm.plot, CO_lm.plot, ncol = 3, rel_widths = c(1,1,2.1))
There isn’t much evidence for a decline in PM2.5, but somewhat more compelling evidence for NO2. We can take a closer look at the estimates:
summary(NO2_lm.stan) ## ## Model Info: ## function: stan_lm ## family: gaussian [identity] ## formula: Concentration ~ Date ## algorithm: sampling ## sample: 4000 (posterior sample size) ## priors: see help('prior_summary') ## observations: 2465 ## predictors: 2 ## ## Estimates: ## mean sd 10% 50% 90% ## (Intercept) 293.7 23.1 263.9 294.1 323.0 ## Date 0.0 0.0 0.0 0.0 0.0 ## sigma 11.3 0.2 11.1 11.3 11.5 ## log-fit_ratio -0.1 0.0 -0.2 -0.1 -0.1 ## R2 0.1 0.0 0.1 0.1 0.1 ## ## Fit Diagnostics: ## mean sd 10% 50% 90% ## mean_PPD 27.0 0.3 26.5 27.0 27.4 ## ## The mean_ppd is the sample average posterior predictive distribution of the outcome variable (for details see help('summary.stanreg')). ## ## MCMC diagnostics ## mcse Rhat n_eff ## (Intercept) 0.9 1.0 671 ## Date 0.0 1.0 671 ## sigma 0.0 1.0 1064 ## log-fit_ratio 0.0 1.0 698 ## R2 0.0 1.0 605 ## mean_PPD 0.0 1.0 3765 ## log-posterior 0.1 1.0 533 ## ## For each parameter, mcse is Monte Carlo standard error, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence Rhat=1). NO2_lm.stan$coefficients ## (Intercept) Date ## 294.07867583 -0.01465859
Here in the case of our NO2 model, the slightly negative log-fit ratio indicates a good fit.
But a simple linear model here is not necessarily the best way to look at these data. Just on the side of the data, there is some heteroskedacity that we might be concerned about. But more importantly, a linear model does not match our actual hypothesis, which is that the pandemic shutdown would have a noticeable effect. A linear model would be better at determining whether there had been a steady decline since the beginning of 2019, but that is not our hypothesis. What we really want to test is whether the “intervention” of the pandemic changed air pollutant concentrations.
To do that, we can look across the various months of April, for example, and see whether April 2020 exceeds the normal variance you’d expect between years.
# Carbon monoxide CO_violin.plot<-ggplot(data=nyc.dat[nyc.dat$Month == 4 & nyc.dat$Year < 2021 & nyc.dat$AQS_PARAMETER_DESC == "Carbon monoxide",])+ geom_violin(aes(x = factor(Year), y = Concentration, fill = Pandemic))+ geom_jitter(aes(x = factor(Year), y = Concentration, color = Pandemic), width = 0.1, alpha = 0.5)+ theme_minimal()+ theme(axis.title.y = element_blank(), legend.position = "None")+ labs(x = "Carbon monoxide")+ ggtitle("")+ scale_fill_manual(values = wes_palette("Darjeeling1")[c(5,1)])+ scale_color_manual(values = wes_palette("Zissou1")[c(3,4)]) # Nitrogen dioxide NO2_violin.plot<-ggplot(data = nyc.dat[nyc.dat$Month == 4 & nyc.dat$Year < 2021 & nyc.dat$AQS_PARAMETER_DESC == "Nitrogen dioxide (NO2)",])+ geom_violin(aes(x = factor(Year), y = Concentration, fill = Pandemic))+ geom_jitter(aes(x = factor(Year), y = Concentration, color = Pandemic), width = 0.1, alpha = 0.5)+ theme_minimal()+ theme(axis.title.y = element_blank(), legend.position = "None")+ labs(x = "Nitrogen dioxide")+ ggtitle("")+ scale_fill_manual(values = wes_palette("Darjeeling1")[c(5,1)])+ scale_color_manual(values = wes_palette("Zissou1")[c(3,4)]) # PM 2.5 PM2_5_violin.plot<-ggplot(data = nyc.dat[nyc.dat$Month == 4 & nyc.dat$Year < 2021 & nyc.dat$AQS_PARAMETER_DESC == "PM2.5 - Local Conditions",])+ geom_violin(aes(x = factor(Year), y = Concentration, fill = Pandemic))+ geom_jitter(aes(x = factor(Year), y = Concentration, color = Pandemic), width = 0.1, alpha = 0.5)+ theme_minimal()+ theme(legend.position = "None")+ ggtitle("April Concentrations")+ labs(x = "PM 2.5")+ scale_fill_manual(values = wes_palette("Darjeeling1")[c(5,1)])+ scale_color_manual(values = wes_palette("Zissou1")[c(3,4)]) # Plot together cowplot::plot_grid(PM2_5_violin.plot, NO2_violin.plot, CO_violin.plot, ncol = 3, rel_widths = c(0.33, 0.33, 0.33))
When viewed this way - comparing Aprils in different years, we can see that April 2020 showed some difference, but it’s not clear in each case whether that difference is substantially different enough from normal that we can think about attributing causation to it. So to answer the question that we’re interested in, we can either do a t-test in a frequentist framework, or a similar approach in a Bayesian framework.
Rather than using Stan here, I’ll implement this model in JAGS, which requires more explicit description of the model. JAGS and RMarkdown don’t get along when creating models, so the model parameterization file is included separately and called here to evaluate.
Now we’ve run the model, and we can have a look at the posterior estimates. In particular here, we’re interested in the estimate for Beta, which is our slope in a linear context. Negative values will tell us that there’s a negative relationship (i.e., that April 2020 did go down) and positive values the opposite.
# Now we can work with the model output g_df<-as.data.frame(g$BUGSoutput$sims.matrix) # First we'll record the credible intervals b_CI<-data.frame(Pollutant = c("CO","CO","NO2","NO2", "PM2.5", "PM2.5"), CI = c(quantile(g_df$beta_CO, 0.025), quantile(g_df$beta_CO, 0.975), quantile(g_df$beta_NO2, 0.025), quantile(g_df$beta_NO2, 0.975), quantile(g_df$beta_PM2_5, 0.025), quantile(g_df$beta_PM2_5, 0.975)), Type = c("Lower","Upper","Lower","Upper","Lower","Upper")) # And then make a new dataframe in which we record all the # posterior estimates that fall within the CIs bCIarea<-data.frame(beta_CO = g_df$beta_CO[g_df$beta_CO > b_CI$CI[1] & g_df$beta_CO < b_CI$CI[2]], beta_NO2 = g_df$beta_NO2[g_df$beta_NO2 > b_CI$CI[3] & g_df$beta_NO2 < b_CI$CI[4]], beta_PM2_5 = g_df$beta_PM2_5[g_df$beta_PM2_5 > b_CI$CI[5] & g_df$beta_PM2_5 < b_CI$CI[6]]) # And then we can plot the posterior estimates for our beta parameter CO_beta.plot<-ggplot(data = g_df, aes(x = beta_CO))+ geom_density(fill = wes_palette("GrandBudapest2")[1], color = wes_palette("GrandBudapest2")[1])+ geom_density(data = bCIarea, fill = wes_palette("GrandBudapest2")[2], color = wes_palette("GrandBudapest2")[2])+ theme_minimal()+ theme(axis.title.y = element_blank())+ labs(x = "Beta CO")+ ggtitle(" ") NO2_beta.plot<-ggplot(data = g_df, aes(x = beta_NO2))+ geom_density(fill = wes_palette("GrandBudapest2")[1], color = wes_palette("GrandBudapest2")[1])+ geom_density(data = bCIarea, fill = wes_palette("GrandBudapest2")[2], color = wes_palette("GrandBudapest2")[2])+ theme_minimal()+ theme(axis.title.y = element_blank())+ labs(x = "Beta NO2")+ ggtitle(" ") PM2_5_beta.plot<-ggplot(data = g_df, aes(x = beta_PM2_5))+ geom_density(fill = wes_palette("GrandBudapest2")[1], color = wes_palette("GrandBudapest2")[1])+ geom_density(data = bCIarea, fill = wes_palette("GrandBudapest2")[2], color = wes_palette("GrandBudapest2")[2])+ theme_minimal()+ labs(x = "Beta PM 2.5", y = "Density")+ ggtitle("Posterior estimates, Beta") # Plot together cowplot::plot_grid(PM2_5_beta.plot, NO2_beta.plot, CO_beta.plot, ncol = 3, rel_widths = c(0.33, 0.33, 0.33))
# Table for CIs knitr::kable(b_CI, caption = "95% Credible Intervals")
95% Credible Intervals Pollutant CI Type CO -0.1166686 Lower CO -0.0678272 Upper NO2 -12.1731875 Lower NO2 -6.9667455 Upper PM2.5 -1.4556021 Lower PM2.5 0.3466315 Upper
Based on our posterior estimates of Beta here, we can see that for PM 2.5, our 95% credible intervals overlap substantially with 0. Areas outside the credible intervals here are pink, while those within are blue. In Bayesian paradigm, this essentially means that there was no significant difference between Aprils prior to Covid and April 2020.
On the other hand, both NO2 and CO show significant drops in concentration for that April.
So at this point, we’ve assessed that concentrations were unusually low in April 2020 for two of our pollutants. Our next step, attempting to understand the causes of this drop, requires more particular knowledge of the typical sources of CO and NO2 in the city. A variety of publicly-available data, including data from the City of New York could help here, but the craft of determining plausible explanations is not always served by merely data-mining as many potential covariates as possible. As previously mentioned, fireworks may have played a role in the lack of a drop in PM 2.5, but the specifics of emissions of each pollutant require a better understanding of systems involved.
Without exploring the potential atmospheric science involved here, we can still compare to other large cities to get a sense for the patter at a larger scale.
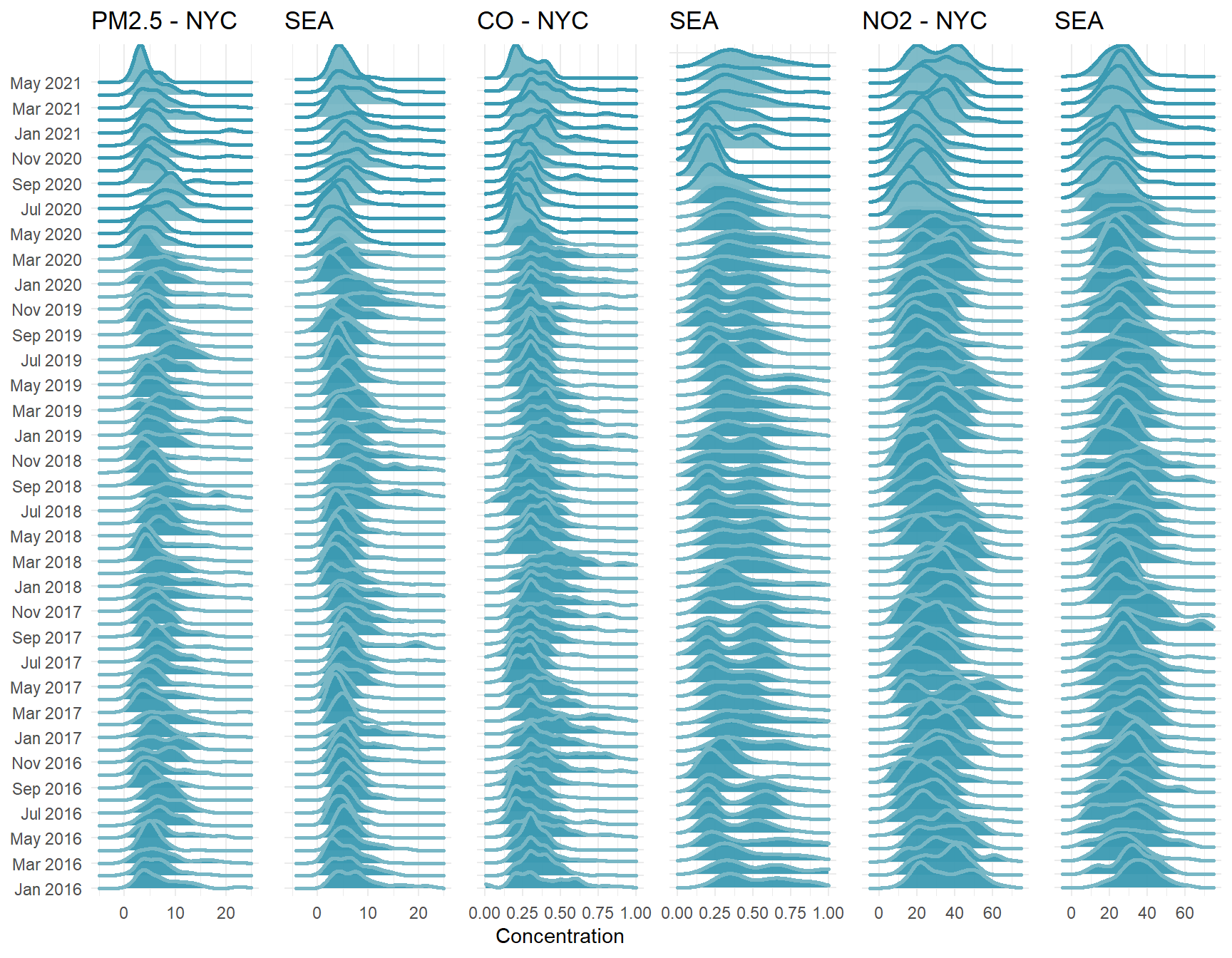
I’ve pulled in data from the EPA’s monitoring stations in Seattle as well. I’ll omit the code as it’s the duplicate of what we’ve done above (but is available in the .RMD), and we can have a look at the comparison with our joy plots again.
# PM 2.5 Plot SEA_joy.pm25<-ggplot(SEA.dat[SEA.dat$AQS_PARAMETER_DESC == pol_types[1],], aes(x = Concentration, y = factor(RunningtextMonth)))+ geom_joy(scale = 3, alpha = 0.95, size = 1, aes(fill = Pandemic, color = Pandemic))+ labs(x = "")+ lims(x = c(-5,25))+ ggtitle("PM 2.5")+ scale_color_manual(values = wes_palette("Zissou1")[c(2,1)])+ scale_fill_manual(values = wes_palette("Zissou1")[c(1,2)])+ scale_y_discrete(breaks = levels(nyc.dat$RunningtextMonth)[c(T,F)])+ theme_minimal()+ theme(legend.position = "None", axis.title.y = element_blank()) # Carbon monoxide plot SEA_joy.CO<-ggplot(SEA.dat[SEA.dat$AQS_PARAMETER_DESC == pol_types[3],], aes(x = Concentration, y = factor(RunningtextMonth)))+ geom_joy(scale = 3, alpha = 0.95, size = 1, aes(fill = Pandemic, color = Pandemic))+ labs(x = " ", y = "")+ lims(x = c(0,1))+ ggtitle(pol_types[3])+ scale_color_manual(values = wes_palette("Zissou1")[c(2,1)])+ scale_fill_manual(values = wes_palette("Zissou1")[c(1,2)])+ theme_minimal()+ theme(legend.position = "None", axis.text.y = element_blank(), axis.title.y = element_blank()) # Nitrogen dioxide plot SEA_joy.NO2<-ggplot(SEA.dat[SEA.dat$AQS_PARAMETER_DESC == pol_types[4],], aes(x = Concentration, y = factor(RunningtextMonth)))+ geom_joy(scale = 3, alpha = 0.95, size = 1, aes(fill = Pandemic, color = Pandemic))+ labs(x = "", y = "")+ lims(x = c(-5,75))+ ggtitle("Nitrogen dioxide")+ scale_color_manual(values = wes_palette("Zissou1")[c(2,1)])+ scale_fill_manual( values = wes_palette("Zissou1")[c(1,2)])+ theme_minimal()+ theme(legend.position = "None", axis.title.y = element_blank(), axis.text.y = element_blank()) # Combine plots - NYC + SEA cowplot::plot_grid(joy.pm25+ggtitle("PM2.5 - NYC"), SEA_joy.pm25+ggtitle("SEA")+ theme(axis.text.y = element_blank()), joy.CO+ggtitle("CO - NYC"), SEA_joy.CO+ggtitle("SEA"), joy.NO2+ggtitle("NO2 - NYC"), SEA_joy.NO2+ggtitle("SEA"), ncol = 6, rel_widths = c(1.4,1,1,1,1,1))
At a broad scale, we see similar patterns. A decline in spring that includes some of the lowest readings recorded of PM2.5 and CO. Perhaps more interesting however, and certainly more noticeable now that we’re looking across two cities, is the decrease in variance for each pollutant as the pandemic shutdown begins. The Seattle area more than NYC has quite variable readings in each month, with broad distributions, but as shutdown begins (March 23 in Seattle), distribution flattens quite a bit. In the model we produced before, we were primarily interested in the mean value for April in each year, but we could also model the variance as well in a hierarchical modelling framework.
Exploring a dataset such as this means understanding the nature of the data it contains - what does each variable mean?, how are observations collected?, where is there independence or not? However there must always be a stage in which creating a useful model relies on domain expertise and conversations with the relevant experts. In this case I would be wary to go much farther without bringing in an atmospheric scientist and having discussions with those who pay attention to drivers of pollutant concentration.
Download the files at Github
It's a Dissertation Defense!
On May 5 I defended my dissertation! Watch here to learn about penguins, seals, exploration, AI, and data science for ecology! Jump ahead to 5:25 when we get started!
Gantarctica
Heading to gantarctica
A year and a half or so ago, considering how poor I am at updating the Lynch Lab’s Instagram (@thelynchlab), I started tooling around with the idea of using a GAN - or Generative Adversarial Network - to just make fake pictures of Antarctica that could be fed into the account. It seemed like a perfect outgrowth of a lot of the work we do, combining Antarctic fieldwork and ecology with high-performance computing and artificial intelligence.
GANs are pretty neat and a little terrifying. A few years ago, NVIDIA (the company that makes graphics cards) did a project in which they used a GAN to generate headshots of fake celebrities. Basically they fed a bunch of pictures of real celebrities into an AI model, which figured out how to make faces that an average person could easily believe were real celebrities.
The process works more or less like this: A generator makes fake data. To start, it doesn’t really know how to fake whatever you’re trying to make. But it’s trying to fool a discriminator. So every time it makes some fake data - in the previous case a fake celebrity face - the discriminator takes a look and has to decide whether it’s real or fake, based on all the data it’s seen (real celebrity photos). Both generator and discriminator learn from this process, but the eventual outcome is that the generator gets so good it can’t really tell what’s real and what’s not. It’s often compared to an art forger and a cop, but this is an imperfect analogy.
In my case, I have a whole library of images of Antarctica that I’ve taken in my 8 years in the field. Gazillions of pictures of penguins, of seals, of people, and of land- and seascapes. It seemed like it shouldn’t be a hard problem, especially since, to my eye, Antarctic landscapes are fairly simple. There’s a much more limited color palette than you’d find in a lot of places, with a lot of white, grey, and a limited selection of blues. Even the sunsets take on a particular yellowish cast.
But for someone like me, with a working understanding of AI but whose skills primarily lie broadly across other disciplines, building up a GAN is not a task for an afternoon. And, with a variety of more pressing projects, it ended up on the back burner.
Recently I came across RunwayML, a web app that is a front end for a pretty expansive number of machine learning architectures. It has StyleGAN - the GAN used for celebrity faces - built in and a really easy interface for uploading training data, training a model, and generating images.
The trick for many image-based machine learning applications is to provide good training data, and to start I decided to take an expansive view. I have a lot of Antarctic images that fall into the landscape category.
PHOTO
So I gathered the images that were explicitly of landscapes along with images of icebergs, of penguins in front of landscapes, of people in landscapes and added them into the training set. Then I pulled in images I’d sourced for another project (grabbed from Google Images and Bing) and photos from other lab members. In it all went. The model expects square images and Runway provides a pre-processing step to crop. There are a few options - you can individually crop, do a center crop, take a random crop, or leave uncropped (in which case they will be squeezed into a square shape). I decided to let it do a random crop, since it was late and I didn’t feel like spending a couple of hours cropping what was now a 750-image training set.
I set up the training process to train for 3000 steps, the default, and let it work.
3000 steps of training - the images get a little more Antarctica-like
The outcome was underwhelming. The images definitely had the feel of Antarctica, but were really poor quality. Plus there were some pretty big problems. It was clear that it had picked up on the people, penguins, and ships present in some of the training data to think that it was okay to put strange, dark shapes in the images. One had a strange wizard-like shape in it. Several had black blobs that might have been inspired by penguins.
Is it a penguin?
I had a couple of options here. I could continue training the model - it obviously needed it since many of the textures were very rough - and hope that it overcame the biggest problems that were there (i.e. that it would give up on penguins and wizards). Or I could start over and more carefully craft the training set. I chose the latter. I pulled out all the pictures in which the landscape was not the main focus (like pictures of penguins with a landscape in the background), plus all the photos that were only of icebergs, photos with ships, photos of people, photos where the horizon wasn’t visible, and ended up with a good set of images basically all showing just mountains, islands, ice, water, and snow.
But my worry was that the training set was too small now. More training data is always better and I was worried it wouldn’t be able to learn textures well if there weren’t enough examples of all the ways that rock or ice can look. So I headed out to the internet.
Using a web-crawler script, I crawled Flickr for all the images tagged “Antarctica” from a variety of areas frequented by tourists. It was a rough pass and I certainly didn’t get all of them, but I managed to pull over a thousand extra images down. Of course with a search that broad, many of those images had the same problems as the original set, so I removed all the pictures that didn’t meet the strict landscape criteria - or which couldn’t be cropped so that they met them.
Then the crop. I worried that part of my problem was the random crop - if I want the GAN to make landscape images, I have to show it only images that are good landscapes. If some of the random crops were only of the sky or water, that would be part of what the GAN would learn. So I manually cropped each of the 1120 images in the training set, choosing a square crop that contained the elements of a decent landscape photograph. Then back to training.
I chose the same 3000 steps so I could monitor the process. This first pass yielded better images - no strange dark blobs - but still had texture issues.
The unifying feature in my mind is improbable geology. There are images of mountains apparently suspended in the air, or large rock formations hanging off of cliffs with seemingly no concern for gravity. More work to be done. The color palette clearly had been ironed out - there are now greens or reds in the images - and the sky in most images contains fairly convincing clouds. Water is challenging - lots of repeated texture or very coarse and unconvincing wave patterns. I continued training for another 1000 steps.
At 4000 steps there’s clear improvement. In small form, there are a numbr of reasonably convincing images. There are some wildly unconvincing ones however, with the same issues of mountains suspended in the air, and strange leaning rocks like before. The water texture has improved quite a bit, but the texture of beaches is still very difficult. I think one of the challenges here is that he cloud ceiling is often quite low in Antarctica, which means that mountains often are swallowed by cloud - cloud that is a somewhat similar color and texture to snow. And sometimes the peaks of those mountains poke up through the clouds again, so it’s understandable that features somewhat like that would pass into the generated images. Still, it can clearly improve.
Onward to 5000 steps.
With 5000 steps we’re getting some quite convincing images. Certainly in small I could believe them. There are some clearly noticeable issue though. Water textures are much better but are still unconvincing in some cases. Most noticeable (now that the water makes some sense) are the reflections. This is the kind of thing that stands out very clearly to a human eye, but is less obvious to a computer. Notice how the reflections don’t really mirror the image perfectly (and some times not at all). There remain a few images that are wildly improbable, such as one (shown above) in which the sea seems to part and flow up the face of a cliff.
Onward to 6000 steps.
At 6000, the same problems exist but they’re less bad. Water is improving. The structure of mountains is very very believable. Icebergs though… The model is clearly a little confused about how icebergs can have strange shapes but mountains should not. I’m not confident it will learn that one before we start overfitting. I’m pretty happy with this one but think there’s some work left. Onward to 7000
I did not expect 7000 steps to be almost universally worse. Of the 50 images I generated, most were nonsensical.
For the next pass, I’ll add 2000 steps to get us to 9000 and see what’s become of it.